Novel Options in GI Diagnostics: DNA Detection of Gut Microbiota
David M. Brady, ND, DC, CCN, DACBN
The population of the microbiota of the human gastrointestinal (GI) tract is widely diverse and complex, with a high population density. All major groups of organisms are represented. While predominately bacteria, a variety of protozoa are also present. In the colon there are more than 1011 bacterial cells per gram and more than 400 different species. These bacterial cells outnumber host cells by at least a factor of 10. This microbial population has important influences on host physiological, nutritional and immunological processes. In fact, this biomass should more rightly be considered a rapidly adapting, renewable organ with considerable metabolic activity and significant influence on human health. Consequently, there is renewed and growing interest in identifying the types and activities of these gut microbes.
The normal, healthy balance in microbiota provides colonization resistance to pathogens. Since anaerobes comprise more than 95% of these organisms, their analysis is of prime importance. Gut microbes might also stimulate immune responses to prevent conditions such as intestinal dysbiosis. Intestinal dysbiosis may be defined as a state of disordered microbial ecology that causes disease. Specifically, the concept of dysbiosis rests on the assumption that patterns of intestinal flora, specifically overgrowth of some microorganisms found commonly in intestinal flora, have an impact on human health. Symptoms and conditions thought to be caused or complicated by dysbiosis include inflammatory bowel diseases, inflammatory or autoimmune disorders, food allergy, atopic eczema, unexplained fatigue, arthritis, mental/emotional disorders in children and adults, malnutrition and breast and colon cancer.
Difficulties in Accurately Assessing Microbiota Content
Most studies of microbiota in the GI tract have used fecal samples. These do not necessarily represent the populations along the entire GI tract from stomach to rectum. Conditions and species can alter greatly along this tract, and generally run from lower to higher population densities. Fecal samples most appropriately represent organisms growing in the colon. In addition, >98% of fecal bacteria will not grow in oxygen. Therefore, standard fecal culture techniques miss the majority of organisms present.
Conventional Techniques vs. New Technologies
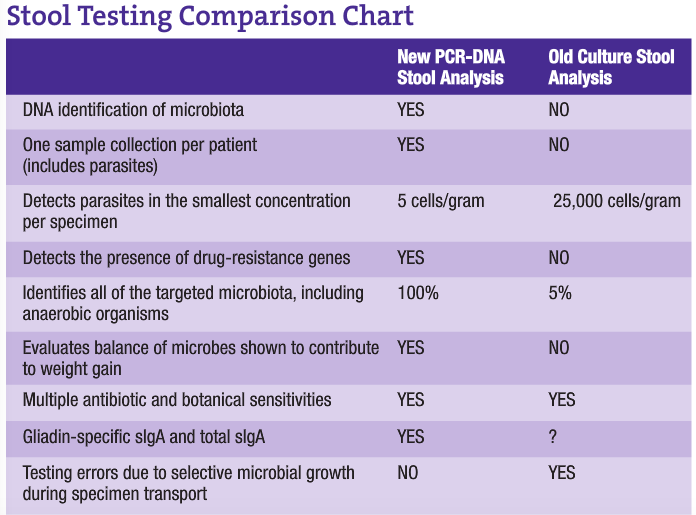
A major problematic issue with present stool analysis procedures is that of transport. Since analysis is culture dependent, sample collection must be done using nutrient broth containers to maintain microbial viability. This allows continued growth of species during transport and until the sample is actually plated out for culture. This growth allows for a significant change in the balance of microbes present, since some species will more actively grow at the expense of others. DNA analysis eliminates this problem by placing the specimen in formalin vials for transport. This immediately kills all organisms, freezing the exact balance present at the time of collection. Since this method is only looking for the genes of microbiota, living specimens are not necessary. This allows the clinician to develop the most appropriate therapy based on the patient’s true gut microbiota, resulting in better clinical results.
Polymerase Chain Reaction (PCR)
One of the most important and profound contributions to molecular biology is the advent of PCR, a DNA- and RNA-based technology that enables us to detect a single genome of an infectious agent in any body fluid with improved accuracy and sensitivity. PCR does not depend on the ability of an organism to grow in culture. Furthermore, PCR is fast, sensitive and capable of copying a single DNA sequence of a viable or non-viable cell over a billion times within three to five hours. The sensitivity of the PCR test is also based on the fact that PCR methodology requires only one to five cells for detection, whereas a positive culture requires an inoculum equivalent to about 1,000-5,000 cells, making PCR the most sensitive detection method available (approximately 1,000 times more sensitive).
Advantages of PCR amplifications of target microbial DNA for organism detection over traditional culture techniques are many:
- Ability to detect nonviable organisms that are not retrievable by culture-based methods
- Ability to detect and identify organisms that cannot be cultured or are extremely difficult to grow (e.g., anaerobes)
- More rapid detection and identification of organisms that grow slowly (e.g., mycobacteria and fungi)
- Ability to detect previously unknown organisms directly in clinical specimens by using broad-range DNA primers
- Ability to quantitate infectious organisms’ burden in patient specimens for better clinical responsiveness.
Parasitology
Parasitology is yet another field of microbiology to be greatly improved with genetic molecular technologies. Parasitic infections are a major cause of nonviral diarrhea, even in developed countries. Classically, parasites have been identified by microscopy and enzyme immunoassays. In recent studies, molecular techniques have proven to be more sensitive and specific than classic laboratory methods. Because Giardia cysts are shed sporadically and the number may vary from day to day, laboratories have adopted multiple stool collections to help increase identification rates for all parasite examinations. And, even with the advent of antigen detection systems, there has long been uncertainty in diagnosis when no ova or parasites are found. Due to the nearly 100% sensitivity and specificity of DNA analysis combined with the need for very low amounts of genomic DNA (as low as 2.5 cells per gram), the previously long specimen collection process, laborious and technically challenging microscopy and resulting delays in reporting have been alleviated. With PCR technology, only one fecal sample is needed for 100% sensitivity and specificity in parasitology examinations.
Intestinal Microbiota Associated With Obesity
There are two predominant bacterial groups in the human GI tract, Bacteroidetes and Firmicutes. Recent research has discovered a relationship between the balance of these groups and obesity. GI bacteria have evolved, in a sense, independently from the host organism, in some cases performing functions that the host consequently has not had to develop and evolve itself. One of these functions is the breakdown of dietary polysaccharides for conversion into energy.
Firmicutes bacteria, which include Bacillus, Clostridium and Lactobacillus species, are very efficient at metabolizing plant polysaccharides into monosaccharides and short-chain fatty acids. These can then be absorbed by the gut and converted to more complex lipids in the liver. In addition, this group secretes a compound that results in increased activity of lipoprotein lipase in adipocytes, resulting in enhanced storage of these lipids. The Bacteroidetes group, which includes Bacteroides and Prevotella species, are not as efficient in this function. Consequently, the balance of these two groups, it has been found, can significantly affect the accumulation of fat stores in the body. While obesity ultimately is caused by excess caloric intake, differences in gut microbial ecology may be an important component of energy homeostasis. In effect, obese individuals may have populations of microbiota that force a more efficient extraction and storage of energy than lean individuals possessing a different balance of microbiota.
This concept of microbiota being linked to obesity raises some interesting possibilities. Studies are ongoing to explore this relationship. The use of specific diets or pre- and probiotic therapies may be able to significantly affect microbial balances that influence fat storage. The ability to assess the balance of these “fat bugs” in humans, as can be done with PCR-based testing of the microbiota, will potentially be an important advance in contributing to the resolution of a significant public health issue, namely obesity.
DNA analysis technology allows for a significant advancement in the understanding of how GI tract microbiota affect human health. It improves patient care by giving clinicians greater options and more tools in treating patients. The increased speed of analysis and improved accuracy makes this the preferred method of stool analysis.
 David M. Brady, ND, DC, CCN, DACBN is a licensed ND, a board-certified Clinical Nutritionist and a Doctor of Chiropractic. He is presently director of the Human Nutrition Institute and an associate professor of clinical sciences at the University of Bridgeport in Connecticut. Dr. Brady also maintains a private practice at The Center for the Healing Arts in Orange, Conn., where he specializes in functional and metabolic medicine.
David M. Brady, ND, DC, CCN, DACBN is a licensed ND, a board-certified Clinical Nutritionist and a Doctor of Chiropractic. He is presently director of the Human Nutrition Institute and an associate professor of clinical sciences at the University of Bridgeport in Connecticut. Dr. Brady also maintains a private practice at The Center for the Healing Arts in Orange, Conn., where he specializes in functional and metabolic medicine.
References
Mackie RI et al: Developmental microbial ecology of the neonatal gastrointestinal tract, Am J Clin Nutr May;69(5):1035S-1045S, 1999.
Hawrelak JA, Myers SP: The causes of intestinal dysbiosis: a review, Altern Med Rev Jun;9(2):180-197, 2004.
Galland L, Barrie S: Intestinal dysbiosis and the causes of diseases, J Advancement Med 6:67-82, 1993.
Savage DC: Microbial ecology of the gastrointestinal tract, Annu Rev Microbiol 31:107-133, 1977.
Berg RD: The indigenous gastrointestinal microflora, Trends Microbiol Nov;4(11):430-435, 1996.
O’Sullivan DJ: Methods of analysis of the intestinal microflora. In: Tannock GW (ed). Probiotics: a critical review. Wymondham, 1999, Horizon Scientific Press, pp. 23-44.
Tannock GW: Analysis of the intestinal microflora: a renaissance, Antonie Van Leeuwenhoek Jul-Nov;76(1-4):265-278, 1999.
Finegold S et al: Normal indigenous intestinal flora. New York, 1983, Academic Press.
Dutta S et al: Sensitivity and performance characteristics of a direct PCR with stool samples in comparison to conventional techniques for diagnosis of Shigella and enteroinvasive Escherichia coli infection in children with acute diarrhoea in Calcutta, India, J Med Microbiol Aug;50(8):667-674, 2001.
Amann RI et al: Phylogenetic identification and in situ detection of individual microbial cells without cultivation, Microbiol Rev Mar;59(1):143-169, 1995.
Wilson KH, Blitchington RB: Human colonic biota studied by ribosomal DNA sequence analysis, Appl Environ Microbiol Jul;62(7):2273-2278, 1996.
Forbes BA et al: Bailey & Scott’s diagnostic microbiology (10th ed). St. Louis, 1998, Mosby.
Verweij JJ et al: Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR, J Clin Microbiol Mar;42(3):1220-1223, 2004.
Ghosh S et al: PCR detection of Giardia lamblia in stool: targeting intergenic spacer region of multicopy rRNA gene, Mol Cell Probes Jun;14(3):181-189, 2000.
Morgan UM et al: Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens: clinical trial, J Clin Microbiol Apr;36(4):995-998, 1998.
Bergeron MG, Ouellette M: Preventing antibiotic resistance through rapid genotypic identification of bacteria and of their antibiotic resistance genes in the clinical microbiology laboratory, J Clin Microbiol, Aug;36(8):2169-2172, 1998.
Martineau F et al: Multiplex PCR assays for the detection of clinically relevant antibiotic resistance genes in staphylococci isolated from patients infected after cardiac surgery. The ESPRIT Trial, J Antimicrob Chemother Oct;46(4):527-534, 2000.
Martineau F et al: Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis, Antimicrob Agents Chemother, Feb;44(2):231-238, 2000.
Backhed F et al: The gut microbiota as an environmental factor that regulates fat storage, Proc Natl Acad Sci USA Nov 2;101(44):15718-15723, 2004.
Ley RE et al: Obesity alters gut microbial ecology, Proc Natl Acad Sci USA Aug 2;102(31):11070-11075, 2005.
Backhed F et al: Mechanisms underlying the resistance to diet-induced obesity in germ-free mice, Proc Natl Acad Sci USA Jan 16;104(3):979-984, 2007.
Turnbaugh PJ et al: An obesity-associated gut microbiome with increased capacity for energy harvest, Nature Dec 21;444(7122):1027-1031, 2006.